ABSTRACT
Smoking is a risk factor for various disease outcomes and is one of the modifiers of DNA methylation. We aimed to identify smoking-related DNA methylation sites (CpG-sites) and test whether one identified CpG-site is associated with smoking-related traits and pulmonary function. We obtained DNA methylation data of 209 men from the Korean Genome and Epidemiology Study analyzed by Illumina's HumanMethylation450 array. To identify smoking-related DNA methylation sites, epigenome-wide association analysis of smoking status was conducted, adjusting for age, area, current drinking status, and body mass index. We assessed the association between smoking intensity and DNA methylation of cg05951221 (AHRR), the CpG showing the strongest largest difference in DNA methylation among the 5 hypomethylated CpGs in current smokers compared to never smokers. The association between DNA methylation and pulmonary function was examined longitudinally resulting in a positive association between DNA methylation and forced expiratory volume in 1 second/forced vital capacity, regardless of adjustment for smoking status. This suggests that DNA methylation associates with long-term pulmonary function. Our study contributes to explaining the relationship between smoking and pulmonary function via DNA methylation.
-
Keywords: Smoking; DNA methylation; Respiratory function tests; Epigenomics
INTRODUCTION
Smoking is widely acknowledged as a risk factor for morbidity and mortality such as cancer, cardiovascular diseases, and pulmonary diseases, attributing to 11.5% of deaths worldwide [
1]. In Korea, smoking is predicted to be associated with a 1.3-fold increase in total mortality [
2] and the burden of disease due to smoking is reported to be 68% in males and 32% in females [
3]. Despite the adverse effect of smoking on health outcomes, the underlying biological mechanism has not been fully elucidated.
One possible explanation of the mechanism by which smoking increases morbidity is through changes in DNA methylation due to smoking. DNA methylation, the addition of methyl groups (CH3
−) to cytosine residue, is one epigenetic mechanism by which gene expression is altered without change in DNA sequence [
4]. Alteration in DNA methylation can be caused by environmental factors including smoking [
5], drinking [
6], diet [
7], and obesity [
8]. Up until now, several epigenome-wide association studies (EWAS) on smoking were conducted to find differentially methylated probes [
9,
10,
11]. However, only very few studies have analyzed smoking-mediated DNA methylation changes in the Korean population [
12,
13].
We first aimed to discover differentially methylated probes associated with smoking by conducting epigenome-wide association analysis in Korean men. Secondly, we assessed whether smoking-related DNA methylation is also associated with other smoking-related traits such as smoking intensity and time since quitting smoking. Lastly, we investigated whether DNA methylation mediates the association between smoking and pulmonary function in cross-sectional and longitudinal study designs.
MATERIALS AND METHODS
Study design and subjects
This study used data of the Korean Genome and Epidemiology Study Ansan-Ansung (KoGES-ASAS) study. The details of the study procedures are provided elsewhere [
14]. Briefly, the KoGES-ASAS study is a longitudinal community-based study that was initiated in 2001-2002 of 10,030 participants aged 40–69 years and followed-up every two years up to the 7th follow-up. This study evaluated data from 4th follow-up (2009–2010) in which DNA methylation profiles were available and to 6th follow-up (2013–2014) with data of pulmonary function test. Out of 6,660 subjects who participated in the 4th follow-up study, 446 subjects were available with DNA methylation data. We restricted our analysis to male subjects, because of large differences in smoking status between male and female (there was only one current smoker in female). Finally, 209 male subjects with DNA methylation profiling were included in the analyses. All study subjects provided written informed consent, and this study was approved by the Institutional Review Board (IRB) of Korea Centers for Disease Control and Prevention (KBP-2019-041) and the IRB of Korea University (KUIRB-2019-0120-01).
Smoking status was assessed by self-reported lifetime cigarette use in 20 packs (400 cigarettes) at 4th follow-up. Participants were categorized as never smokers (never smoked or smoked less than 20 packs of cigarettes), former smokers (smoked more than 20 packs of cigarettes, but did not smoke at the time of the survey), and current smokers (smoked more than 20 packs of cigarettes and smoke at the time of the survey). To estimate smoking intensity, pack-years were calculated by multiplying the number of packs of cigarettes smoked per day by the number of years smoked. The time since quitting (years) was reported by former smokers. To avoid the risk of misclassification of smoking status and make it more accurate, we reclassified the smoking status by using previously surveyed variables. Those who smoked previously, but responded as “never-smokers” currently were excluded, because their smoking-related traits were not surveyed and can bias the results in any direction.
Pulmonary function test
Pulmonary function was measured using a spirometer (VMAX 2130, SensorMedics, Yorba Linda, CA) following the 1994 American Thoracic Society guidelines [
15]. The forced vital capacity (FVC, L), and forced expiratory volume in 1 second (FEV1, L) were measured and FEV1/FVC was calculated by FEV1 divided by FVC.
For DNA methylation profiling, 446 participants were sampled from KoGES-ASAS study after the quality control of samples having DNA under 100 ng per sample, or with gender discrepancy. DNA methylation was measured using the Illumina Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA, USA) [
16]. Quality control was performed on the DNA methylation data (485,577 CpGs) filtering out the probes with detection p value over 0.05. We additionally excluded probes at the X and Y chromosomes. The DNA methylation level, measured in beta-value was calculated using GenomeStudio Methylation Module Software, ranging from 0 (unmethylated) to 1 (methylated). After all the filtering, a total of 473,004 CpGs remained for epigenome-wide association analysis.
Descriptive statistics of all the variables were reported as mean ± standard error (SE) for continuous variables and as percentages and the number of counts for categorical variables by smoking status in
Table 1. Epigenome-wide association analysis was conducted in R version 3.6.1 using the CpGassoc package [
17] to model smoking status (current vs. never) as the exposure and DNA methylation (beta-value) as the outcome. Each EWAS was tested with age and area (Ansan/Ansung) as covariates in the basic model, and with age, area, current drinking status, and body mass index as covariates in the fully adjusted model. To correct for multiple testing, the Holm step-down Bonferroni (p value < 1.0E-7) and Benjamini-Hochberg method (false discovery rate [FDR] < 0.05) were used to assess significance. The results are presented as a Manhattan plot and quantile-quantile plot in
Figure 1, and details are presented in
Table 2. The associations between DNA methylation and smoking-related traits were visualized using boxplot combined with jitter, or scatter plot with the fitted line in
Figure 2. The association between DNA methylation and pulmonary function was examined in cross-sectional and longitudinal study design. The multiple linear regression or multilevel mixed-effects generalized linear model was applied, respectively. In both models, the fully adjusted model with age, area, current drinking status, and body mass index as covariates was applied. To account for the effect of smoking on pulmonary function, smoking status was additionally adjusted in both analyses. For longitudinal analysis, stratified analysis by smoking status was performed, to investigate whether the mediating effect exists.
Table 1Characteristics of study participants by smoking status
Table 1
|
Characteristics |
Smoking status |
|
Never (n = 40) |
Former (n = 78) |
Current (n = 91) |
p value*
|
|
Age (yr) |
58.2 ± 1.3 |
59.2 ± 0.9 |
58.0 ± 0.8 |
0.583 |
|
Area (Ansan/Ansung %) |
35.0/65.0 |
43.6/56.4 |
44.0/56.0 |
0.598 |
|
Current drinker (%, N) |
24 (60.0) |
59 (75.6) |
76 (44.4) |
0.015 |
|
Body mass index (kg/m2) |
24.2 ± 0.6 |
24.3 ± 0.4 |
23.6 ± 0.3 |
0.292 |
|
Smoking traits |
|
|
|
|
|
Smoking amount (pack-years) |
- |
26.2 ± 2.8 |
30.9 ± 1.8 |
0.143 |
|
Time since quitting (years) |
- |
13.5 ± 1.1 |
- |
- |
|
Pulmonary function |
|
|
|
|
|
FVC (L) |
4.12 ± 0.10 |
4.07 ± 0.08 |
4.09 ± 0.08 |
0.957 |
|
FEV1 (L) |
3.26 ± 0.09 |
3.07 ± 0.08 |
3.07 ± 0.06 |
0.224 |
|
FEV1/FVC |
79.19 ± 0.90b
|
75.03 ± 1.01a
|
75.24 ± 0.92a
|
0.017 |
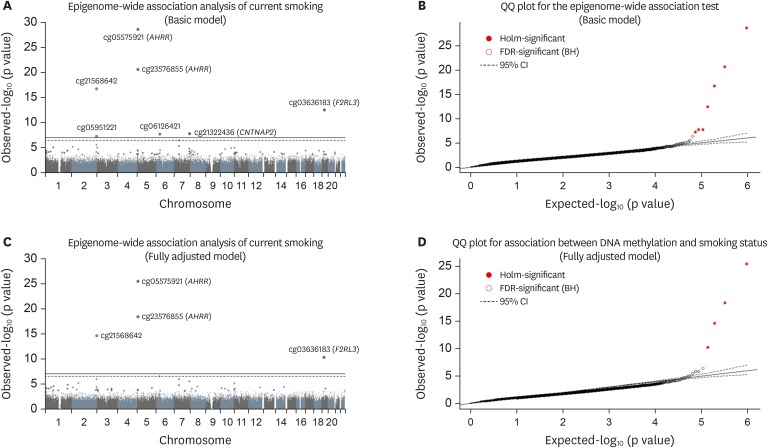
Figure 1Epigenome-wide association analysis of smoking status. (A, C) Manhattan plot showing chromosome location of 22 autosomes (X-asix) and -log10(p-value) for each CpG (Y-axis). The top line indicates Holm step-down Bonferroni threshold (p value < 1.0E-7) and the bottom (dashed) line indicates the Benjamini-Hochberg threshold (FDR < 0.05). A regression model was applied for epigenome-wide association analysis adjusted for age, and area in the basic model. (B, D) Quantile-quantile plot showing observed vs. expected −log10(p value) for association at all CpG sites. A regression model was applied for epigenome-wide association analysis, adjusted for and age, area, drinking status, and body mass index in the fully adjusted model.
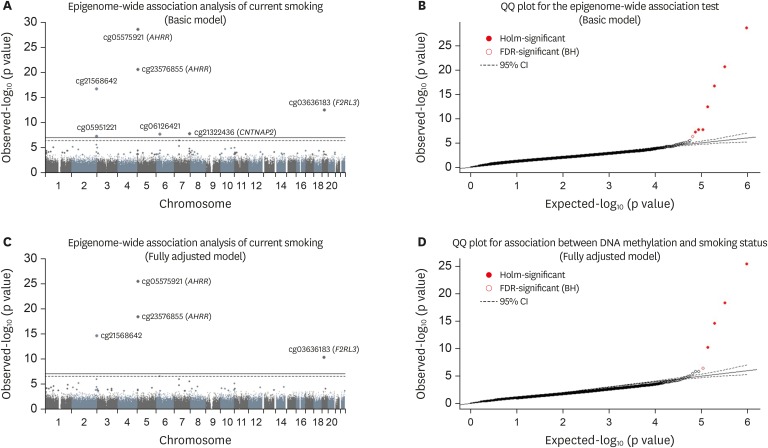
Table 2Difference in methylated CpG sites between current smokers and never smokers (FDR < 0.05)
Table 2
|
Probe ID |
CHR |
Position |
Gene |
Basic model*
|
Fully adjusted model†
|
|
Beta |
SE |
p value |
FDR |
Beta |
SE |
p value |
FDR |
|
cg05575921 |
5 |
373378 |
AHRR
|
−0.206 |
0.014 |
2.19E-29 |
1.03E-23 |
−0.204 |
0.015 |
3.86E-26 |
1.83E-20 |
|
cg23576855 |
5 |
373299 |
AHRR
|
−0.160 |
0.014 |
2.30E-21 |
5.45E-16 |
−0.162 |
0.015 |
4.81E-19 |
1.14E-13 |
|
cg21566642 |
2 |
233284661 |
2q37.1
|
−0.112 |
0.011 |
1.98E-17 |
3.12E-12 |
−0.111 |
0.012 |
2.65E-15 |
4.17E-10 |
|
cg03636183 |
19 |
17000585 |
F2RL3
|
−0.100 |
0.012 |
3.49E-13 |
4.13E-08 |
−0.095 |
0.013 |
5.94E-11 |
7.03E-06 |
|
cg21322436 |
7 |
145812842 |
CNTNAP2
|
−0.042 |
0.007 |
1.84E-08 |
1.60E-03 |
- |
- |
- |
- |
|
cg06126421 |
6 |
30720080 |
6p21.33
|
−0.087 |
0.014 |
2.03E-08 |
1.60E-03 |
−0.064 |
0.013 |
3.87E-07 |
3.66E-02 |
|
cg05951221 |
2 |
233284402 |
2q37.1
|
−0.067 |
0.012 |
5.94E-08 |
4.01E-03 |
- |
- |
- |
- |
|
cg12803068 |
7 |
45002919 |
MYO1G
|
0.072 |
0.014 |
4.75E-07 |
2.81E-02 |
- |
- |
- |
- |
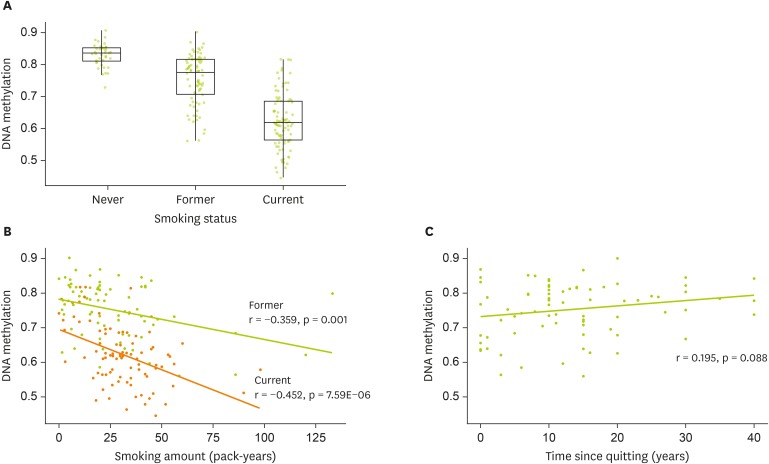
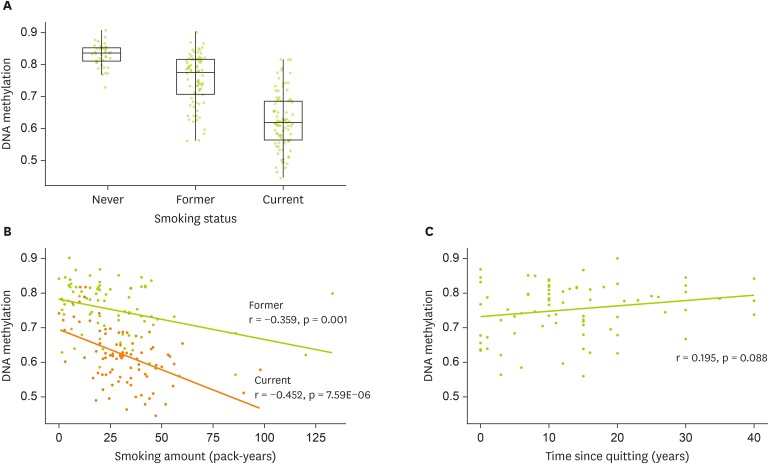
Figure 2DNA methylation at cg05575921 (AHRR) and smoking-related traits (A) Box plot showing the distribution of DNA methylation by smoking status (B) Scatter plot with the fitted line showing the association between DNA methylation and pack-year by smoking status (C) Scatter plot with the fitted line showing the association between DNA methylation and time since quitting smoking.
RESULTS
Characteristics of study participants
The characteristics of study participants by smoking status are shown in
Table 1. Among the 209 participants in our study, 40 were never smokers (19.1%), 78 were former smokers (37.3%), and 91 were current smokers (45.5%). Age, area, and body mass index were not different by smoking status, whereas the percentage of current drinkers was lower in current smokers than never smokers (44.4% vs. 60.0%; p = 0.015). For pulmonary function test results, current smokers and former smokers had lower pulmonary function compared to never smokers when assessed by FEV1/FVC ratio.
We compared DNA methylation profiles between the 91 current smokers and 40 never smokers. The results are presented in
Table 2 and
Figure 1. In the basic model, 7 sites showed differences according to the Holm step-down Bonferroni procedure (p value < 1.0E-7), and 8 sites showed significance by the Benjamini-Hochberg procedure (FDR < 0.05). In the fully adjusted model, 4 sites showed significant differences according to the Holm step-down Bonferroni procedure, and 5 sites showed significance by the Benjamini-Hochberg procedure. Except for cg12803068 (
MYO1G), other significant CpGs were hypomethylated in current smokers compared to never smokers. cg05951221 (
AHRR) showed the largest difference in DNA methylation and was thus selected for further analysis.
Compared to never smokers (0.83 ± 0.01), DNA methylation at cg05951221 was lower in former (0.75 ± 0.01; p = 3.59E-08) and current smokers (0.62 ± 0.01; p = 4.09E-09) (
Figure 2A). Consumption of smoking measured in pack-years was negatively correlated with DNA methylation in both former (r = −0.359; p = 0.001) and current smokers (r = −0.452; p = 7.59E-06) (
Figure 2B). Time since quitting tended to be positively correlated with DNA methylation (r = 0.195; p = 0.088) (
Figure 2C).
We investigated whether DNA methylation is associated with pulmonary function cross-sectionally and longitudinally (
Table 3). In our cross-sectional analysis, DNA methylation was positively associated with FEV1/FVC in the fully adjusted model (β = 9.63; 95% confidence interval [CI], 0.28 to 18.98; p = 0.044), but the association disappeared when additionally adjusted for smoking status. Whereas in the longitudinal analysis, DNA methylation and pulmonary function were positive associated regardless of adjustment for smoking status (β = 14.88; 95% CI, 4.47 to 25.29; p = 0.005). This may indicate that DNA methylation associates with pulmonary function on a long-term basis, rather than on a short-term basis. Stratification according to smoking status revealed that DNA methylation was not associated with FEV1/FVC in current smokers (β = 4.68; 95% CI = −14.50 to 23.85; p = 0.632) or never smokers (β = 19.02; 95% CI, −34.15 to 72.19; p = 0.483). However, a positive association was observed in former smokers (β = 19.96; 95% CI, 2.27 to 37.68; p = 0.027). These results point to the possibility that the effect of current smoking itself on pulmonary function may override the effect of DNA methylation on pulmonary function.
Table 3The association between DNA methylation at cg05575921 and pulmonary function
Table 3
|
Variables |
Fully adjusted model‡
|
Fully adjusted model + smoking status§
|
|
Beta coefficient (95% CI) |
p value |
Beta coefficient (95% CI) |
p value |
|
Cross-sectional analysis*
|
|
|
|
|
|
FVC (L) |
−0.01 (−0.82 to 0.80) |
0.973 |
−0.03 (−1.30 to 1.23) |
0.959 |
|
FEV1 (L) |
0.46 (−0.21 to 1.11) |
0.181 |
0.27 (−0.75 to1.29) |
0.600 |
|
FEV1/FVC |
9.63 (0.28 to 18.98) |
0.044 |
6.57 (−7.27 to 20.41) |
0.350 |
|
Longitudinal analysis†
|
|
|
|
|
|
FVC (L) |
−0.14 (−0.88 to 0.60) |
0.715 |
−0.52 (−1.35 to 0.31) |
0.218 |
|
FEV1 (L) |
0.53 (−0.11 to 1.17) |
0.104 |
0.21 (−0.48 to 0.91) |
0.552 |
|
FEV1/FVC |
14.98 (5.69 to 24.26) |
0.002 |
14.88 (4.47 to 75.29) |
0.005 |
DISCUSSION
We identified 5 CpG-sites associated with smoking status in Korean men. Among these sites, methylation of cg05575921 (AHRR) was most strongly associated with smoking status. Methylation at this site was positively associated with FEV1/FVC ratio at 4-year follow-up, regardless of smoking status.
The effect of smoking on repression of cg05575921 (
AHRR) methylation is universal among races. Prior studies on EWASs of smoking status were mainly conducted in whites or blacks [
9,
10,
18,
19,
20,
21,
22,
23]. In a systematic review of 14 EWAS of smoking, 7 studies were from Europe, 6 investigated blacks, and 1 was performed in Qatar [
18]. Unfortunately, ethnic differences were not investigated in this study. The three most frequently reported CpG-sites were cg05575921 (
AHRR), cg03636183 (
F2RL3), cg19859270 (
CPR15), followed by CpG-sites at
2q37.1 and
6p21.33, which were also found in our analyses. In another meta-analysis of DNA methylation in 15,907 participants of 16 cohorts, 2,623 CpG-sites were identified to be differentially methylated according to smoking status [
23]. The results of this study were similar to previous studies, where cg05575921 on
AHRR showed the strongest significance and largest effect size (18% hypomethylated in current smokers; p = 4.6E-26). In this meta-analysis paper, ancestry-stratified analysis was conducted resulting in differences in the related CpG-sites between whites and blacks. Still the correlation between the effect estimates of CpG-site significant in at least one ancestry and in entire group of two ancestries were highly correlated (r = 0.89). The effect size and significance in the five CpG-sites (cg05575921 at
AHRR, cg23576855 at
AHRR, cg21566642 at
2q37.1, cg03636183 at
F2RL3, and cg06126421 at
6p21.33) in our study were similar to white and black current smokers (e.g. cg05575921 at
AHRR; 18% hypomethylated in European current smokers; 16% hypomethylated in African current smokers). Regarding Asians, 2 previous EWAS of smoking have been performed in the Korean population. The first EWAS was conducted in 2013 using a Korean chronic obstructive pulmonary disease (COPD) cohort, and identified 108 significant probes, in comparison of 31 current and 39 never smokers [
12]. The second EWAS was conducted in 2019 using the Korean Healthy Twin (KHT) cohort and identified 6 CpGs associated with smoking in 190 monozygotic twins (FDR < 0.05) [
13]. Our analysis identified 5 significant CpGs associated with smoking in 131 middle-aged Korean males (91 current smokers vs. 40 never smokers) from the KoGES-ASAS study. Among all three studies, cg05575921 was the most associated CpG-site with smoking status. Therefore, suppressed methylation of cg05575921 by smoking is evident regardless of race.
Of the observed CpG-sites associated with smoking in our study, 5 were also reported in the 2 previous studies performed in Koreans. Two CpG-sites (cg05575921 at
AHRR, and cg03636183 at
F2RL3) were reported in the COPD cohort and three CpG-sites (cg05575921, cg23576855 at
AHRR, and cg21566642 at
2q37.1) were reported of in the KHT cohort study. One CpG-site uniquely discovered in the current study is cg06126421 at
6p21.33 which was marginally associated with smoking status in one of EWAS in Korean population (p = 2.6E-04) [
12]. The difference in study population (i.e. COPD, twin, or general population limited to male subjects), covariates adjusted in the analysis (i.e. COPD status, or whether the cell-type composition was adjusted), and analysis method (i.e. empirical Bayes method using limma, or linear mixed model) may explain the differences in the results.
Our analyses were limited to Korean men due to the low incidence of smoking in Korean women. Unlike other studies with 7-45% of female current smokers [
9,
20,
21,
22], the rate of female current smokers was as low as 0.4% (1 in 237 participants) in our study. Within the aforementioned systemic review of 14 EWAS, two studies included only males, and four included only females due to the inclusion criteria in each cohort. Despite the suggested sex differences in smoking-related methylation profiles [
5], the significance of CpG-sites affected by smoking status and the magnitude of the effect size of the association were not different between the 14 studies. Although the current study cannot confirm the same effect on female smokers, a high possibility exists that the same CpG-sites found to be less methylated in smoking men also have a low methylation rate in smoking women compared to non-smoking women.
The association of DNA methylation with smoking intensity (pack-year) and time since quitting demonstrates that the effect of smoking can be reversed, but the benefit does not appear rapidly. The negative relationship between cg05951221 (
AHRR) DNA methylation and smoking intensity and the positive trend between cg05951221 (
AHRR) DNA methylation and time since quitting are in line with previous EWASs results [
10,
12,
23]. Hypomethylation in the
AHRR region is caused by exposure to polycyclic aromatic hydrocarbons in cigarette smoke [
24]. It has been reported that DNA methylation can be recovered by quitting smoking [
23,
25,
26]. The rapid decline in DNA methylation with increased pack-years in smokers is alleviated in previous smokers, but the increase in DNA methylation after quitting takes decades. This long-term effect of smoking on DNA methylation possibly affects pulmonary function at 4 years follow-up.
Our study is the first to longitudinally address the association between smoking-related CpG-sites and lung function. The effect of smoking-induced DNA methylation on pulmonary function has been studied using mediation analysis [
27,
28,
29,
30] and Mendelian randomization [
29,
30]. The indirect effect suggested a mediating effect of DNA methylation on FEV1 [
27], while another mediation study reported ten CpG-sites mediating FEV1, and five of these CpG-sites were validated in lung tissues [
28]. Others found a possible causal effect of DNA methylation on pulmonary function using both mediation and Mendelian randomization analysis [
29,
30]. However, questions still remained on causality [
31]. In an EWAS of Korean COPD patients and lung function, lung function related CpG-sites has not tested the association with smoking status [
32]. By assessing DNA methylation and lung function over a 4-year period, we add more weight on the causal relationship between low methylation of CpG due to smoking and lung function.
Limitations of this study include the lack of consideration of cell composition and batch effect in performing EWAS. However, this was inevitable as the relevant information was not available. Another limitation includes the inability to replicate our results in another independent dataset. Although false positive results may exist, the differentially methylated CpG sites reported in this study have been reported in previous studies and are well-known for their associations with smoking status.
The strength of this study is that this study is the first to utilize DNA methylation data of 446 subjects profiled in the KoGES-ASAS 4th follow-up study. This DNA methylation profiling was performed in a relatively large sample of the general population. Second, owing to the longitudinal study design, the possible effect of DNA methylation on pulmonary function could be prospectively examined. Therefore, the results can be somewhat free from reverse causality compared to a cross-sectional design. Third, misclassification of smoking status of some participants was considered, and smoking status was re-categorized. Self-reported smoking status may underestimate the true smoking status [
33], which may lead to biased results.
CONCLUSION
We identified 5 CpG sites associated with smoking in epigenome-wide association analysis in Korean men. Our results are in line with previous EWAS conducted in Koreans and other races. The targeted CpG-site cg05575921 (AHRR) was associated with smoking status and amount. Our longitudinal analysis shows that smoking may affect pulmonary function via DNA methylation.
National Research Foundation of Koreahttps://doi.org/10.13039/501100003725NRF-2019R1F1A1063744
NOTES
-
Funding: This study was supported by the Basic Science Research Program through the NRF funded by the Ministry of Science and ICT (NRF-2019R1F1A1063744).
-
Conflict of Interest: The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
This study was conducted with bioresources from National Biobank of Korea, the Centers for Disease Control and Prevention, Republic of Korea (KBP-2019-041).
REFERENCES
- 1. GBD 2015 Tobacco Collaborators. Smoking prevalence and attributable disease burden in 195 countries and territories, 1990–2015: a systematic analysis from the Global Burden of Disease Study 2015. Lancet 2017;389:1885-1906.
- 2. Lee EH, Park SK, Ko KP, Cho IS, Chang SH, Shin HR, Kang D, Yoo KY. Cigarette smoking and mortality in the Korean Multi-center Cancer Cohort (KMCC) study. J Prev Med Public Health 2010;43:151-158.
- 3. Zahra A, Cheong HK, Park JH. Burden of disease attributable to smoking in Korea. Asia Pac J Public Health 2017;29:47-59.
- 4. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23-38.
- 5. Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet 2013;4:132.
- 6. Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kühnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JB, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry 2018;23:422-433.
- 7. Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol 2012;863:359-376.
- 8. Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, Waldenberger M, Peters A, Erdmann J, Hengstenberg C, Cambien F, Goodall AH, Ouwehand WH, Schunkert H, Thompson JR, Spector TD, Gieger C, Trégouët DA, Deloukas P, Samani NJ. DNA methylation and body-mass index: a genome-wide analysis. Lancet 2014;383:1990-1998.
- 9. Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet 2013;22:843-851.
- 10. Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, Strauch K, Waldenberger M, Illig T. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 2013;8:e63812.
- 11. Zhu X, Li J, Deng S, Yu K, Liu X, Deng Q, Sun H, Zhang X, He M, Guo H, Chen W, Yuan J, Zhang B, Kuang D, He X, Bai Y, Han X, Liu B, Li X, Yang L, Jiang H, Zhang Y, Hu J, Cheng L, Luo X, Mei W, Zhou Z, Sun S, Zhang L, Liu C, Guo Y, Zhang Z, Hu FB, Liang L, Wu T. Genome-wide analysis of DNA methylation and cigarette smoking in a chinese population. Environ Health Perspect 2016;124:966-973.
- 12. Lee MK, Hong Y, Kim SY, London SJ, Kim WJ. DNA methylation and smoking in Korean adults: epigenome-wide association study. Clin Epigenetics 2016;8:103.
- 13. Kim EA. DNA methylation changes as an exposure signature of cigarette smoking [master's thesis]. Seoul: Seoul National University; 2019.
- 14. Kim Y, Han BG. KoGES group. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) Consortium. Int J Epidemiol 2017;46:1350.
- 15. American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med 1995;152:1107-1136.
- 16. Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288-295.
- 17. Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics 2012;28:1280-1281.
- 18. Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics 2015;7:113.
- 19. Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, Davey Smith G, Hughes AD, Chaturvedi N, Relton CL. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics 2014;6:4.
- 20. Harlid S, Xu Z, Panduri V, Sandler DP, Taylor JA. CpG sites associated with cigarette smoking: analysis of epigenome-wide data from the Sister Study. Environ Health Perspect 2014;122:673-678.
- 21. Guida F, Sandanger TM, Castagné R, Campanella G, Polidoro S, Palli D, Krogh V, Tumino R, Sacerdote C, Panico S, Severi G, Kyrtopoulos SA, Georgiadis P, Vermeulen RC, Lund E, Vineis P, Chadeau-Hyam M. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet 2015;24:2349-2359.
- 22. Dogan MV, Shields B, Cutrona C, Gao L, Gibbons FX, Simons R, Monick M, Brody GH, Tan K, Beach SR, Philibert RA. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics 2014;15:151.
- 23. Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Smith JA, Brody JA, Dhingra R, Yousefi P, Pankow JS, Kunze S, Shah SH, McRae AF, Lohman K, Sha J, Absher DM, Ferrucci L, Zhao W, Demerath EW, Bressler J, Grove ML, Huan T, Liu C, Mendelson MM, Yao C, Kiel DP, Peters A, Wang-Sattler R, Visscher PM, Wray NR, Starr JM, Ding J, Rodriguez CJ, Wareham NJ, Irvin MR, Zhi D, Barrdahl M, Vineis P, Ambatipudi S, Uitterlinden AG, Hofman A, Schwartz J, Colicino E, Hou L, Vokonas PS, Hernandez DG, Singleton AB, Bandinelli S, Turner ST, Ware EB, Smith AK, Klengel T, Binder EB, Psaty BM, Taylor KD, Gharib SA, Swenson BR, Liang L, DeMeo DL, O'Connor GT, Herceg Z, Ressler KJ, Conneely KN, Sotoodehnia N, Kardia SL, Melzer D, Baccarelli AA, van Meurs JB, Romieu I, Arnett DK, Ong KK, Liu Y, Waldenberger M, Deary IJ, Fornage M, Levy D, London SJ. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 2016;9:436-447.
- 24. Watanabe T, Imoto I, Kosugi Y, Fukuda Y, Mimura J, Fujii Y, Isaka K, Takayama M, Sato A, Inazawa J. Human arylhydrocarbon receptor repressor (AHRR) gene: genomic structure and analysis of polymorphism in endometriosis. J Hum Genet 2001;46:342-346.
- 25. Tsaprouni LG, Yang TP, Bell J, Dick KJ, Kanoni S, Nisbet J, Viñuela A, Grundberg E, Nelson CP, Meduri E, Buil A, Cambien F, Hengstenberg C, Erdmann J, Schunkert H, Goodall AH, Ouwehand WH, Dermitzakis E, Spector TD, Samani NJ, Deloukas P. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 2014;9:1382-1396.
- 26. Philibert R, Hollenbeck N, Andersen E, McElroy S, Wilson S, Vercande K, Beach SR, Osborn T, Gerrard M, Gibbons FX, Wang K. Reversion of AHRR demethylation is a quantitative biomarker of smoking cessation. Front Psychiatry 2016;7:55.
- 27. Barfield R, Shen J, Just AC, Vokonas PS, Schwartz J, Baccarelli AA, VanderWeele TJ, Lin X. Testing for the indirect effect under the null for genome-wide mediation analyses. Genet Epidemiol 2017;41:824-833.
- 28. de Vries M, van der Plaat DA, Nedeljkovic I, Verkaik-Schakel RN, Kooistra W, Amin N, van Duijn CM, Brandsma CA, van Diemen CC, Vonk JM, Marike Boezen H. From blood to lung tissue: effect of cigarette smoke on DNA methylation and lung function. Respir Res 2018;19:212.
- 29. Imboden M, Wielscher M, Rezwan FI, Amaral AF, Schaffner E, Jeong A, Beckmeyer-Borowko A, Harris SE, Starr JM, Deary IJ, Flexeder C, Waldenberger M, Peters A, Schulz H, Chen S, Sunny SK, Karmaus WJ, Jiang Y, Erhart G, Kronenberg F, Arathimos R, Sharp GC, Henderson AJ, Fu Y, Piirilä P, Pietiläinen KH, Ollikainen M, Johansson A, Gyllensten U, de Vries M, van der Plaat DA, de Jong K, Boezen HM, Hall IP, Tobin MD, Jarvelin MR, Holloway JW, Jarvis D, Probst-Hensch NM. Epigenome-wide association study of lung function level and its change. Eur Respir J 2019;54:54.
- 30. Jamieson E, Korologou-Linden R, Wootton RE, Guyatt AL, Battram T, Burrows K, Gaunt TR, Tobin MD, Munafò M, Davey Smith G, Tilling K, Relton C, Richardson TG, Richmond RC. Smoking, DNA methylation, and lung function: a mendelian randomization analysis to investigate causal pathways. Am J Hum Genet 2020;106:315-326.
- 31. London SJ. Methylation, smoking, and reduced lung function. Eur Respir J 2019;54:1900920.
- 32. Lee MK, Hong Y, Kim SY, Kim WJ, London SJ. Epigenome-wide association study of chronic obstructive pulmonary disease and lung function in Koreans. Epigenomics 2017;9:971-984.
- 33. Pérez-Stable EJ, Marín G, Marín BV, Benowitz NL. Misclassification of smoking status by self-reported cigarette consumption. Am Rev Respir Dis 1992;145:53-57.